BIOSS - 3D and 4D Image Analysis for the 4D-Analyser (DFG)
Project Members
- Junior-Prof. Dr. Olaf Ronneberger
- Prof. Dr.-Ing. Thomas Brox
- Prof. Dr.-Ing. Hans Burkhardt
- Margret Keuper
- Henrik Skibbe
- Robert Bensch
- Benjamin Drayer
- Kun Liu
Introduction
Official BIOSS website
Bioss Research Areas
- BIOSS Area A: Intra-cellular Signalling Pathways
- BIOSS Area B: Supra-cellular Signalling Pathways
- BIOSS Area C: Rebuilding and Synthesis
- BIOSS Area D: Development and Technology Platform (Pathway Assembler, 4D-Analyser)
- BIOSS Incubator
Universal Image Analysis (for the BIOSS 4D-Analyser)
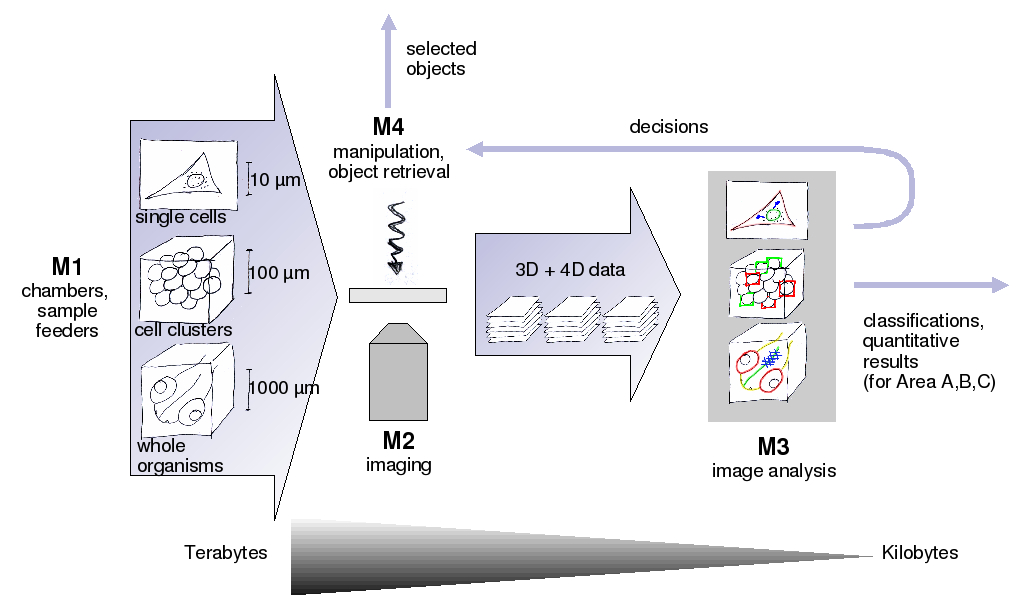
The goal of the project "D2-1 Universal Image Analysis" is to provide universal image analysis modules for the 4D Analyser. These modules will be "plugged together" to extract the biologically relevant quantitative parameters from the huge amount of recorded 3D/4D data, and to provide decisions for an automated manipulation of the experiments. One unique feature of our modules will be the adaption of a module to the specific application just by (interactive) training instead of tedious manual parameter adjustment. Further goals are an integrated, easy to understand, but powerful data and module management, which allows an efficient development for the computer scientists, and at the same time an easy and intuitive usage for the end-user. The goal for all modules is to reach a full automation, such that high-throughput experiments become possible.
The modules are split into four categories:
|
Publications
| [1] |
M. Reisert and H. Burkhardt.
Complex derivative filters.
IEEE Transactions on Image Processing, 17(12):2265-2274, 2008. |
| [2] |
Qing Wang, Olaf Ronneberger, and Hans Burkhardt.
Rotational invariance based on fourier analysis in polar and
spherical coordinates.
IEEE Transactions on Pattern Analysis and Machine Intelligence,
31:1715-1722, 2009. [ .pdf ] |
| [3] |
M. Emmenlauer, O. Ronneberger, A. Ponti, P. Schwarb, A. Griffa, A. Filippi,
R. Nitschke, W. Driever, and H. Burkhardt.
Xuvtools: free, fast and reliable stitching of large 3d datasets.
Journal of Microscopy, 233(1):42-60, 2009. [ .pdf ] |
| [4] |
R. Bensch, O. Ronnerberger, B. Greese, C. Fleck, K. Wester, M. Hülskamp,
and H. Burkhardt.
Image analysis of arabidopsis trichome patterning in 4d confocal
datasets.
In Biomedical Imaging: From Nano to Macro, 2009. ISBI 2009. 6th
IEEE International Symposium on, pages 742-745, 2009. [ .pdf ] |
| [5] |
Maja Temerinac-Ott and Hans Burkhardt.
Multichannel image restoration based on optimization of the
structural similarity index.
In Asilomar Conference on Signals, Systems, and Computers, nov
2009. [ .pdf ] |
| [6] |
Q. Wang, O. Ronneberger, E. Schulze, R. Baumeister, and H. Burkhardt.
Using lateral coupled snakes for modeling the contours of worms.
In Proceedings of the DAGM 2009, LNCS, pages 542-551, Jena,
Germany, 2009. Springer. |
| [7] |
Margret Keuper, Jan Padeken, Patrick Heun, Hans Burkhardt, and Olaf
Ronneberger.
A 3d active surface model for the accurate segmentation of
drosophila schneider cell nuclei and nucleoli.
In Advances in Visual Computing. Proceedings of the 5th
International Symposium on Visual Computing, ISVC 2009, Part I, volume 5875
of Lecture Notes in Computer Science, pages 865-874. Springer, 2009. [ http | .pdf ] |
| [8] |
Maja Temerinac, Margret Keuper, and Hans Burkhardt.
Evaluation of a new point clouds registration method based on group
averaging features.
In Proceedings of 20th International Conference on Pattern
Recognition (ICPR 2010), Istanbul, Turkey, 2010.
(accepted). |
| [9] |
M. Keuper, T. Schmidt, J. Padeken, P. Heun, K. Palme, H. Burkhardt, and
O. Ronneberger.
3d deformable surfaces with locally self-adjusting parameters - a
robust method to determine cell nucleus shapes.
In Proceedings of the 20th International Conference on Pattern
Recognition (ICPR 2010), Istanbul, Turkey, 2010.
accepted. |
| [10] |
M. Keuper, J. Padeken, P. Heun, H. Burkhardt, and O. Ronneberger.
Mean shift gradient vector flow: A robust external force field for 3d
active surfaces.
In Proceedings of the 20th International Conference on Pattern
Recognition (ICPR 2010), Istanbul, Turkey, 2010.
accepted. |
| [11] |
M Schlachter, M Reisert, C Herz, F Schluermann, S Lassmann, M Werner,
H Burkhardt, and O Ronneberger.
Harmonic filters for 3d multi-channel data: Rotation invariant
detection of mitoses in colorectal cancer.
Medical Imaging, IEEE Transactions on, PP(nn):nn, 2010.
accepted for Publication. |
| [12] |
Dorothee Saur, Olaf Ronneberger, Dorothee Kummerer, Irina Mader, Cornelius
Weiller, and Stefan Kloppel.
Early functional magnetic resonance imaging activations predict
language outcome after stroke.
Brain, 133(4):1252-1264, 2010. |
| [13] |
Margret Keuper, Robert Bensch, Karsten Voigt, Alexander Dovzhenko, Klaus Palme,
Hans Burkhardt, and Olaf Ronneberger.
Semi-supervised learning of edge filters for volumetric image
segmentation.
In Proceedings of the DAGM 2010, LNCS, page nn, Darmstadt,
Germany, 2010. Springer.
to appear. |
| [14] |
Soeren Lienkamp, Athina Ganner, Christopher Boehlke, Thorsten Schmidt,
Sebastian J. Arnold, Tobias Schäfer, Daniel Romaker, Julia Schuler,
Sylvia Hoff, Christian Powelske, Annekathrin Eifler, Corinna Krönig, Axel
Bullerkotte, Roland Nitschke, E. Wolfgang Kuehn, Emily Kim, Hans Burkhardt,
Thomas Brox, Olaf Ronneberger, Joachim Gloy, and Gerd Walz.
Inversin relays frizzled-8 signals to promote proximal pronephros
development.
Proceedings of the National Academy of Sciences,
107(47):20388-20393, 2010. |


