U-Net: Convolutional Networks for Biomedical Image Segmentation
The u-net is convolutional network architecture for fast and precise segmentation of images. Up to now it has outperformed the prior best method (a sliding-window convolutional network) on the ISBI challenge for segmentation of neuronal structures in electron microscopic stacks. It has won the Grand Challenge for Computer-Automated Detection of Caries in Bitewing Radiography at ISBI 2015, and it has won the Cell Tracking Challenge at ISBI 2015 on the two most challenging transmitted light microscopy categories (Phase contrast and DIC microscopy) by a large margin (See also our annoucement).
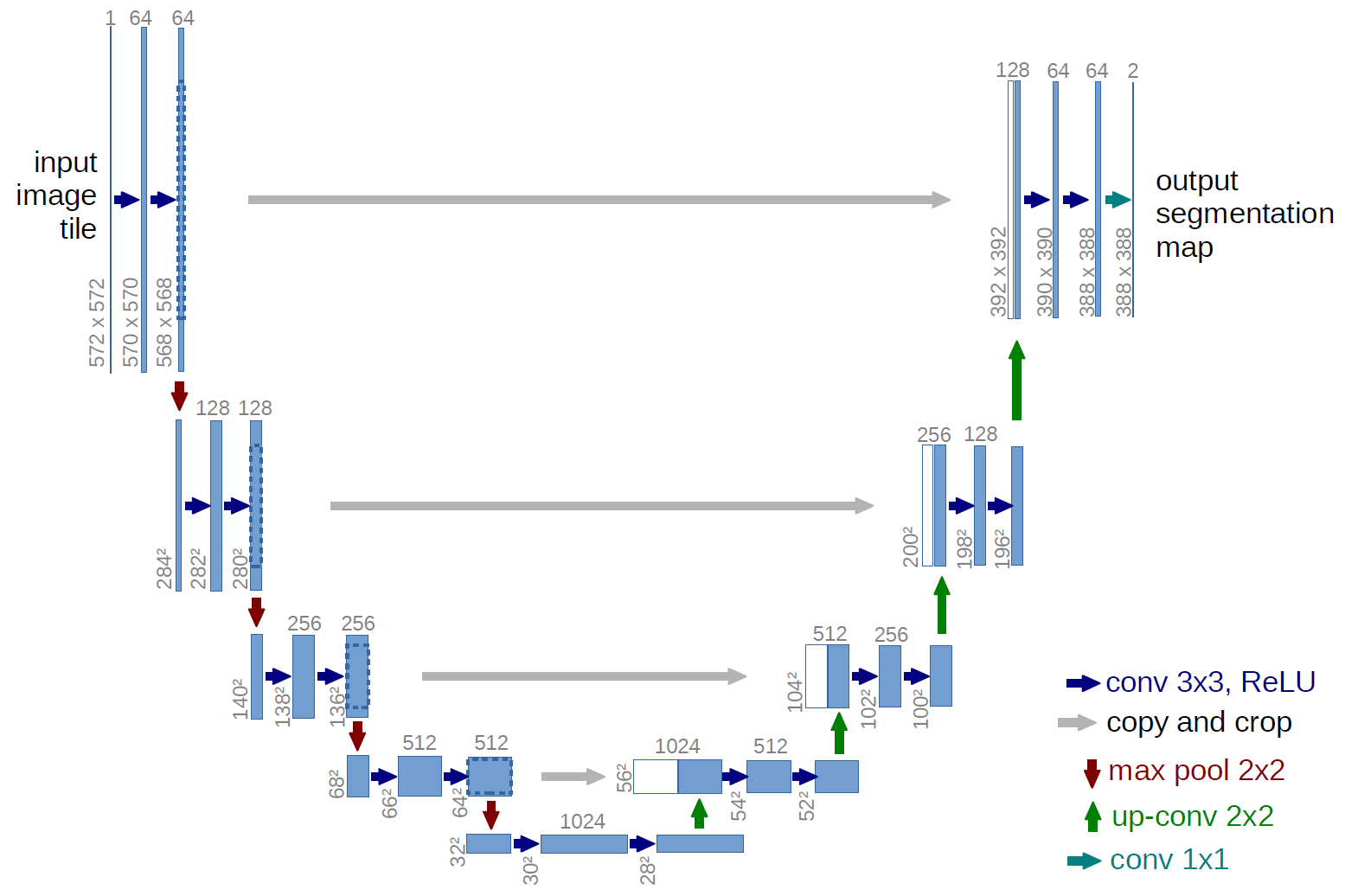
Article describing U-net

|
U-Net: Convolutional Networks for Biomedical Image Segmentation
 Publisher's Link Publisher's Link
Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol.9351: 234--241, 2015, available at arXiv:1505.04597 [cs.CV]
|
5 Minute Teaser Presentation of the U-net
Download video: u-net-teaser.mp4 (68MB)
Download
We provide the u-net for download in the following archive: u-net-release-2015-10-02.tar.gz (185MB). It contains the ready trained network, the source code, the matlab binaries of the modified caffe network, all essential third party libraries, the matlab-interface for overlap-tile segmentation and a greedy tracking algorithm used for our submission for the ISBI cell tracking challenge 2015. Everything is compiled and tested only on Ubuntu Linux 14.04 and Matlab 2014b (x64)
To apply the segmentation and the tracking to the images in "PhC-C2DH-U373/01" simply run the shell script
./segmentAndTrack.sh
The resulting segmentation masks will be written to "PhC-C2DH-U373/01_RES"
If you do not have a CUDA-capable GPU or your GPU is smaller than mine, edit segmentAndTrack.sh accordingly (see there for documentation). If you have any questions, you may contact me at ronneber@informatik.uni-freiburg.de, but be aware that I can not provide any support.
Olaf Ronneberger


