The iRoCS pipeline - a walk-through example
Prerequisites
To follow the tutorial, please download rootTip_short.h5, and the SVM models (nucleusDetector_svmModel.h5, epidermisLabelling_svmModel.h5 and layerAssignment_svmModel.h5).
Import a raw dataset
Choose Channel->Import Channels and select rootTip_short.h5 on your local hard drive. A window will pop up showing the internal file structure of the selected hdf5 file. You should see the root group and a subgroup t0. Unroll the group t0 by clicking on the arrow to the left of the group name. You should see the group contents, in the case of rootTip_short.h5 the dataset channel0, containing a 3D recording of a DAPI stained root tip. Select the dataset channel0 and click OK. A progress dialog will pop up while the channel is loading. Don't worry if your application blocks for a short period of time, it will recover as soon as the dataset has been loaded. After loading you will see a black and white volume in the orthoview. Check the Normalize checkbox in the channel control widget to adapt the gray value range to the display range.
Navigation within the dataset
You can navigate through the volume by dragging the cross-hair or moving the mouse wheel while your mouse is over one of the view panels. If you hold the Ctrl key pressed while moving the mouse wheel, the view is zoomed. You can move the splitters in between the view panels to fit it to your needs, even hide complete views, if you only need one view of a huge data set. On the right a second dock panel is available, showing information on the data under the cursor, namely the cursor position in micrometers and voxel coordinates relative to the upper left frontal data set corner and the data value under the cursor (if multiple channels are loaded the values of each channel are shown separated by whitespaces).
Adapting the channel view with the Channel Control widget
The channel control widget on the right shows new options, that will be described now in detail.
Channel
When multiple channels are loaded you can select the active channel. All settings within the Channel Control widget apply to the active channel only. You can remove the current channel from the workspace by clicking on the delete button right to the channel selector.
Channel Name
You can rename the channel by editing this text field. When saving the project, this name corresponds to the dataset/group name, that will be created in the output hdf5 file.
Alpha (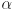 )]
)]
When moving this slider or editing the corresponding spin box you change the weight of the current channel within the orthoview rendering. The orthoview shows the weighted sum of the voxels of all loaded channels in their corresponding colors:
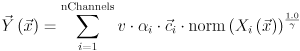
Visible ( )
)
You can toggle, whether a channel is rendered or not using this switch.
Color (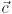 )
)
You can select the channel color by either setting the values in the spinboxes, that correspond to the red, green and blue components of the RGB colorspace or you can click on the palette button on the right to choose from the system palette.
Gamma (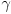 )
)
You can increase the contrast by gamma correcting the gray values. Greater
![]() -values spread the low grayvalue range which often leads to
a better visual impression.
-values spread the low grayvalue range which often leads to
a better visual impression.
Normalize (norm)
When enabling normalization the gray value range will be normalized to show the darkest gray value as black and the brightest as white (min-max-normalization).
Show Exposure Problems
When this switch is toggled on, the darkest and brightest gray values occuring in the channel will be replaced by blue (under-exposure) and red (over-exposure) respectively.
Nucleus detection
Choose Plugins->01 Detect Nuclei to detect the nuclei. A dialog will pop up asking for some information:
- Data Channel
- With this combo box you can select the data channel to detect nuclei in. Currently only one data channel is loaded and you should see its name in the combo box.
- Annotation channel
- The plugin will create an annotation channel to store the detected nuclei. You can leave it at its default value for the tutorial. The selected name can be changed after running the plugin using the channel control widget
- Feature File
- Prior to nucleus detection the voxel-wise gray value features are computed and stored in the file you give here. Make sure that enough hard-disk space is available to store the features. It will need approximately 400 times the size of the original dataset!
- Model File
- Choose the file nucleusDetector_svmModel.h5 you downloaded.
After setting up the plugin, you can start the nucleus detection process by clicking OK. Get yourself a big cup of coffee until the detection process is finished. Two new channels should have been generated after detection, an annotation channel named as given in the dialog containing nucleus markers (indicated by spheres in the orthoview) and a data channel /annotation/detector/decisionValues that shows the SVM decision values resulting from the voxelwise classification. It is colored red and shows peaks at the probable nucleus center positions.
Epidermis labelling
Choose Plugins->02 Label Epidermis to label the epidermal nuclei. A dialog similar to the one before will pop up:
- Data Channel
- With this combo box you can select the data channel to classify the nuclei in. Select the original DAPI channel if not already selected.
- Annotation channel
- Select the annotation channel that has been created during the detection process. Currently only one annotation channel should be available, thus the selection should point to the correct channel.
- Feature File
- Choose the feature file, that has been created during the detection process to reuse the already stored features. That will speed up the classification a lot. If you removed the feature file generated during the detection process it will be regenerated with the given name.
- Model File
- Choose the file epidermisLabelling_svmModel.h5.
Start the pre-classification by clicking OK.
Selecting the QC
Create a new annotation channel by choosing Channel->New Annotation Channel. A dialog will pop up allowing you to choose between different marker types, namely point markers (which suffice for the choice of the quiescent center position), sphere markers which add the freedom to select a radius and nucleus markers containing all relevant meta-data to describe the nuclei within iRoCS. Select Point as marker type and click OK. Now navigate through the dataset until you reach the z-plane containing the quiescent center and select it by clicking once onto the QC center position. A small cross appears, marking QC. You can change the position by editing the values of the position attribute in the Marker Control widget on the right or by dragging the cross with the mouse. The label is not important and can remain -1. If you accidentally clicked more than once, you can remove markers by either right clicking near them in the orthoview or by selecting Remove Marker from the Operation combo box in the channel control widget and left clicking the markers you want to remove.
Attaching iRoCS to the dataset
Choose Plugins->03 Attach iRoCS to estimate the root axis and the intrinsic positions of all nucleus candidates. Again a dialog will ask for the necessary information:
- Marker Channel
- Select the channel containing the nucleus candidates from the detection phase (Default: /annotation/detector).
- QC Channel
- Select the annotation channel you created in the previous step.
- Feature File
- Select the feature file you created during the detection process. The voxelwise continuous iRoCS coordinates will be added as new features.
- Feature Group
- Leave this text field at its default value.
Now click OK to fit the continuous iRoCS to your dataset. At first glance you won't notice a difference. But each nucleus marker in the annotation channel contains the intrinsic coordinates as set of attributes now. You can verify this by selecting the annotation channel containing your nucleus candidates, selecting Edit marker as Operation and selecting one marker by clicking on it in the orthoview. The Marker control widget will appear and show the marker meta-data. When toggling the Show all checkbox, you get additional read-only information, including the iRoCS coordinates in the attributes QC distance, Radial Distance and Phi.
Layer assignment
Choose Plugins->04 Assign Layers to start the layer assignment that uses appearance and positional information. The following parameters need to be provided:
- Data Channel
- Select the original DAPI channel.
- Annotation Channel
- Select the annotation channel containing the nucleus candidates.
- Feature File
- Select the feature file you created during the detection process and extended during the Attach iRoCS step.
- Model File
- Choose the file layerAssignment_svmModel.h5.
Start the layer assignment by clicking OK.
Manual post-processing (if desired)
After running the pipeline most of the nucleus candidates are correctly identified, but there are still obvious mis-classifications, that can be corrected manually if highly accurate results are needed especially on small populations. For this select the annotation channel containing the nucleus candidates and select as operation Edit Marker. Select a marker to edit by clicking it in the orthoview. The Marker control widget will be updated showing the meta information of the selected marker. Now you can edit the fields in the Marker Control Widget. If you want to add missing markers or want to remove false positives, select Add Marker or Delete Marker as operation. In Add Marker-Mode, a right click also removes the marker clicked on. If you want to change the labels of many markers in sequence setting them to the same label you can choose Label Marker as operation. The selected marker presets will be applied to any marker clicked. E.g. assume you want to label all nuclei of layer epidermis, then you chose as operation Label Marker, enable the presets for Label, set it to 2 (Epidermis) and disable all other presets. That way each click on a marker only changes its label attribute to the preset value 2 leaving all other meta-information untouched. After manual editing you need to run the plugin 03 - Attach iRoCS again to update the coordinate system and assign the new coordinates to all markers. You should not reclassify after manual editing, because your manually provided labels will be overwritten! If the coordinate system could not be attached successfully due to too bad epidermis labelling you can also edit the markers to initialize the RANSAC fit.


