Check, if the software runs on your Linux
cd ~/vibez_tutorial
./vibez_elastic_registration -h
You should get a detailed description of all parameters like that:
vibez_elastic_registration -- Elastic Registration of Two Volumetric Images
---------------------------------------------------------------------------
Elastic registration of two volumetric images using and the locally
normalized cross correlataion as similarity measure and fast combinatortial
optimization based on the approach of Glocker et al. 2008
Parameters:
fixedfile=... (default: reference_embryo_2011_05_23.h5)
file name of HDF5 file containing the fixed image.
fixedgroup=... (default: /step4b/rigidToAtlasManual/long)
HDF5 group, that contains the data sets (channels) for the
fixed image
channelname1=... (default: channel0)
HDF5 data set name of first channel (usually the nucleus
channel as anatomic refernce). Used for moving and fixed
file
channelname2=... (default: )
HDF5 data set name of an additional channel (usually the
gene expression pattern). Will be transformed identically
like the first channel, but not used for findeing the
correspondences. Used for moving and fixed file
movingfile=... (default: )
file name of HDF5 file containing the moving image. If
omitted, the global input file (specified with -i) is used
movinggroup=... (default: /step4/fused/)
HDF5 group, that contains the data sets (channels) for the
moving image
fixedmaskfile=... (default: )
(optional) file name of HDF5 file containing a mask for the
fixed image.
fixedmaskdataset=... (default: )
HDF5 data set name (full path), of the data set containing
a mask for the fixed image. Must have the same shape and
resolution like the fixed image
movingmaskfile=... (default: )
(optional) file name of HDF5 file containing a mask for the
moving image.
movingmaskdataset=... (default: )
HDF5 data set name (full path), of the data set containing
a mask for the moving image. Must have the same shape and
resolution like the moving image
outfilename=... (default: )
file name of output file. If omitted, the global input file
(specified with -i) is used
outgroup=... (default: /step7/elasticToAtlasAuto)
HDF5 group, for the channels of the registered image, and
the deformation field.
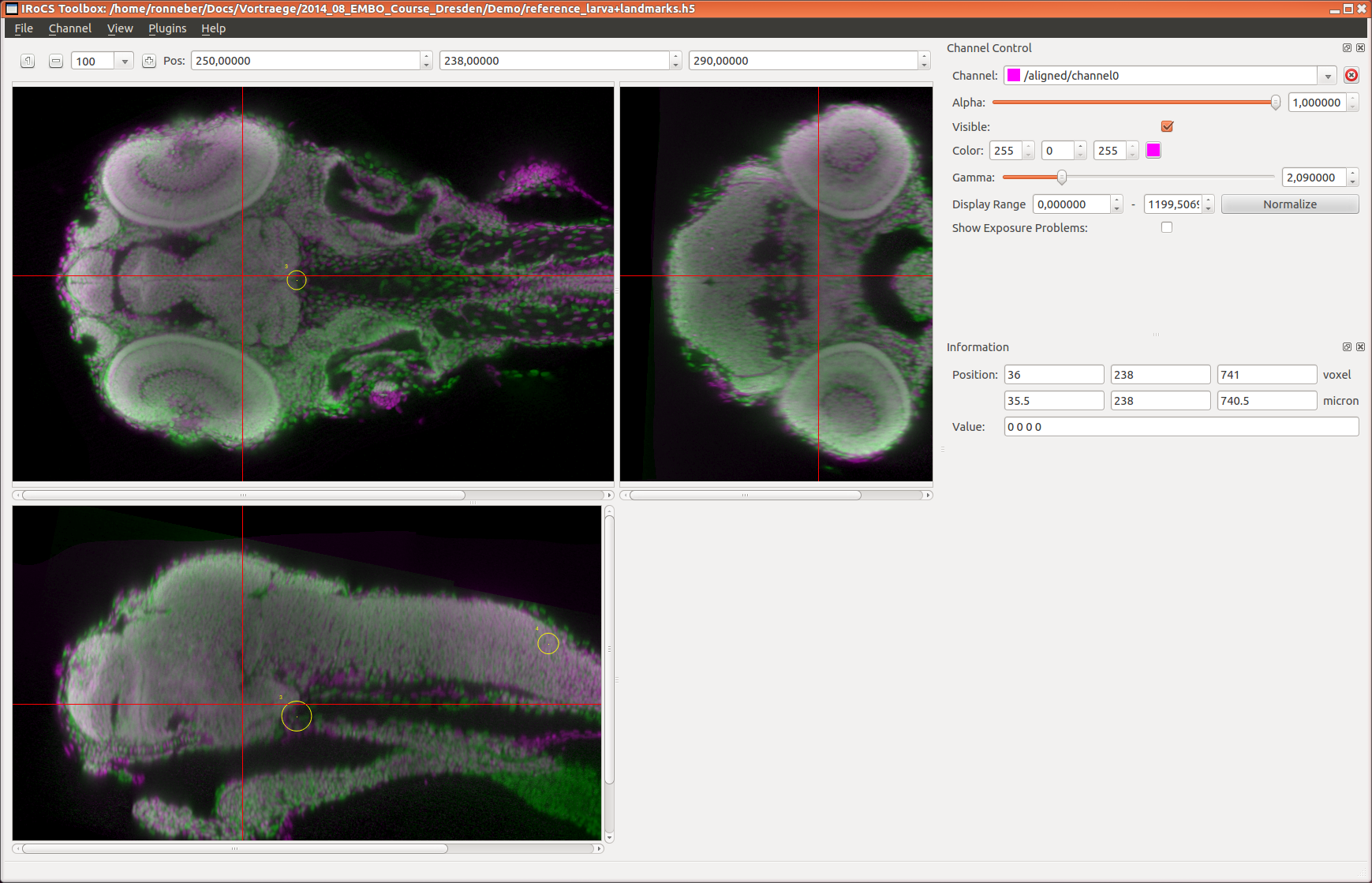
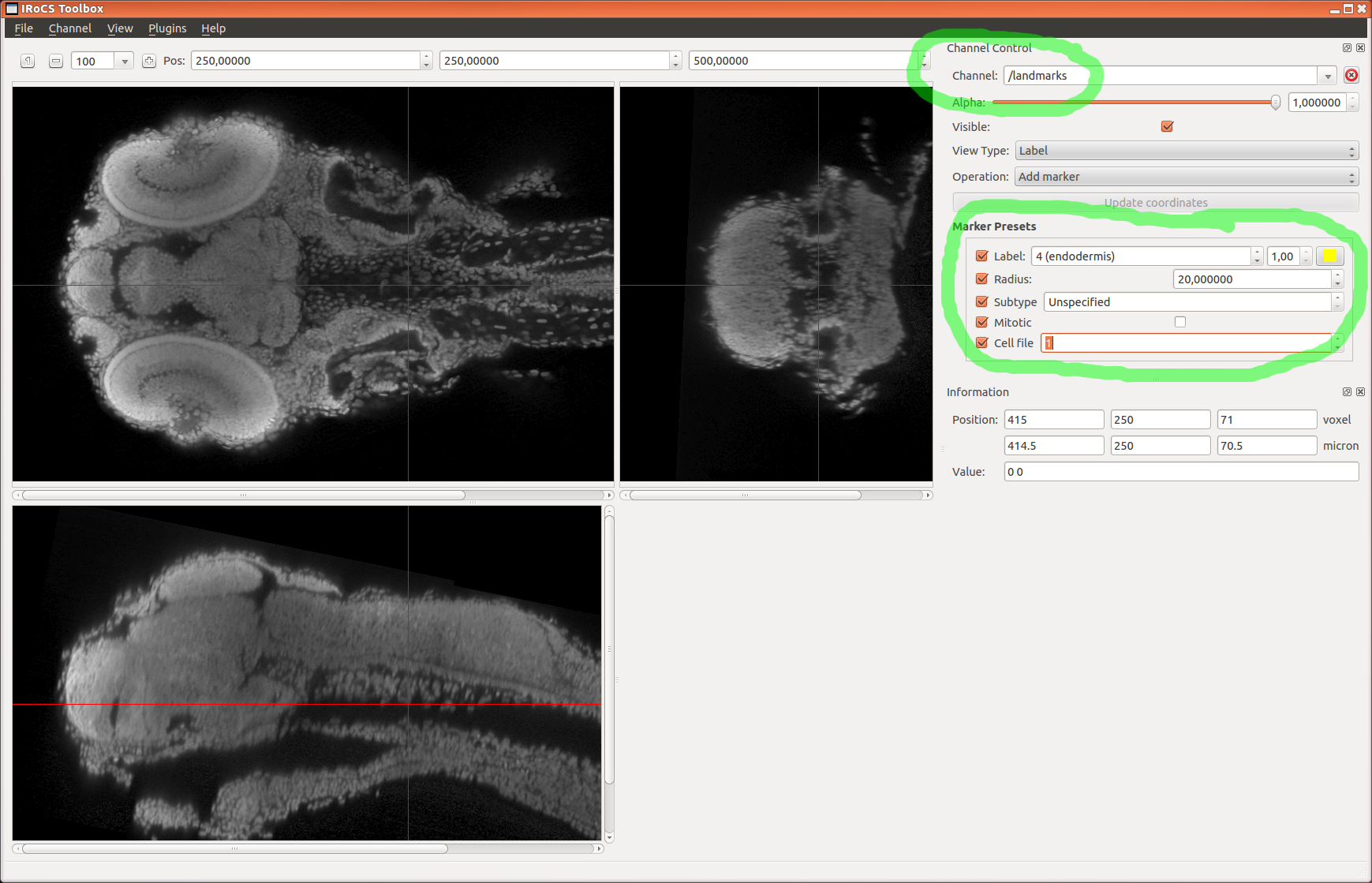
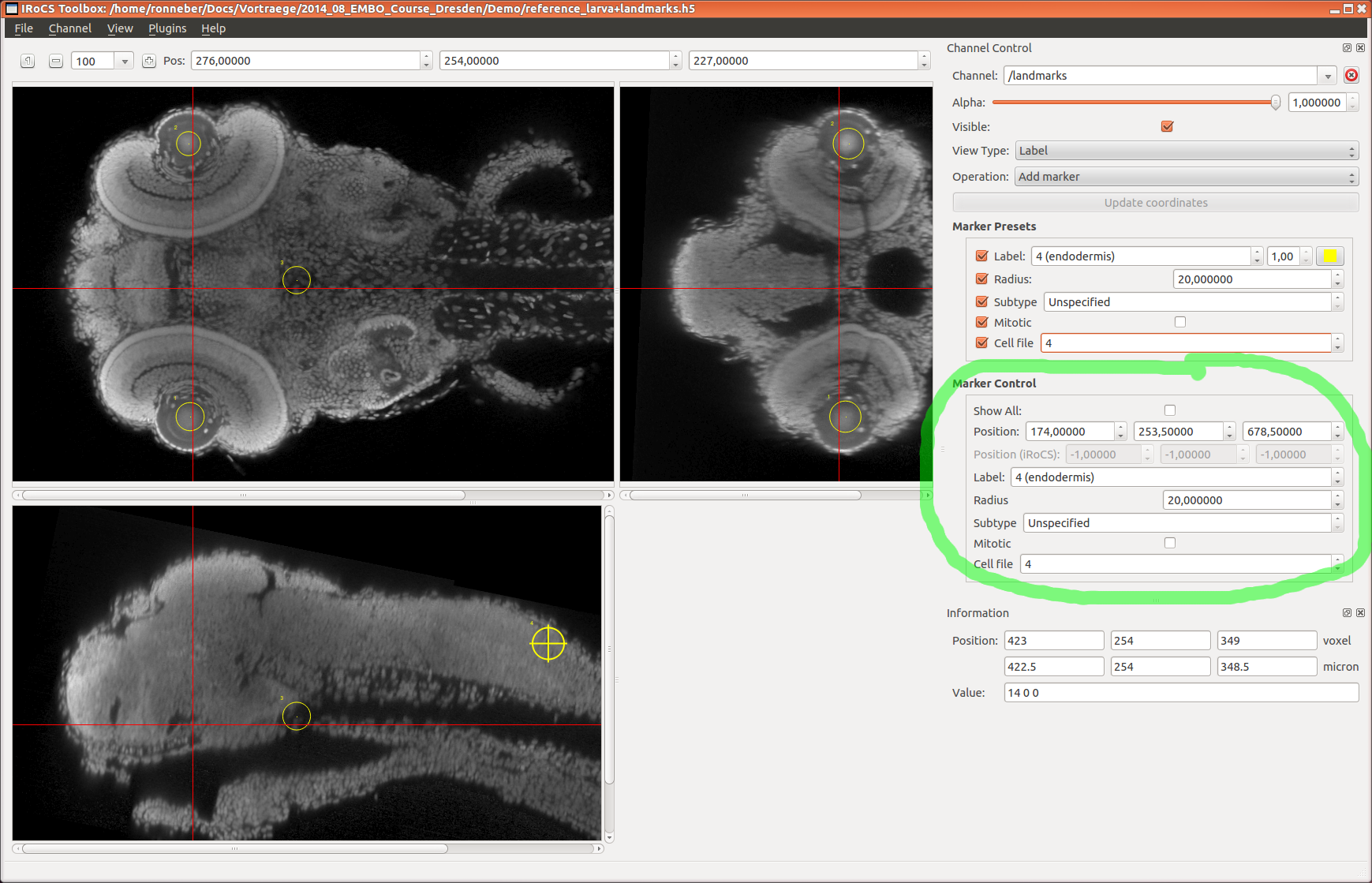
fixedlandmarks=... (default: )
(optional) HDF5 group, that contains the landmarks in the
fixed image (have to be stored in the datasets
'position_um' and 'label')
movinglandmarks=... (default: )
(optional) HDF5 group, that contains the landmarks in the
moving image (have to be stored in the datasets
'position_um' and 'label')
landmarks_label_dataset=... (default: continuity)
(optional) HDF5 data set name, that contains the landmarks
labels ('label' if you used iRoCS point or sphere markers,
or 'continuity' if you used nucleaus markers and cell file
labels). Fallback to 'label', if 'continuity' data set does
not exist
use_landmarks_label=... (default: )
(optional) specify a subset of landmarks that shall by used
for the initial transformation. e.g.
use_landmarks_label=0,5,6,11,12,13,14,15
landmarktrafo=... (default: umeyama)
specifies, how to use the landmarks for the initial
transformation. 'umeyama' finds the best matching rigid
transformation (translation and rotation). 'TPS' uses thin
plate splines to obtain an elastic transformation field,
that maps exactly each pair of landmarks.
fixedToMovingMat=... (default: )
(optional) 4x4 affine transformation matrix for the initial
transfroamtion. All units are in micrometer, the coordinate
origin is at the upper left frontal corner (the first
element of the volumetric data set).The matrix has top map
a position in the fixed image (fixpos) to the corresponding
position in the moving image (movpos), such that movpos =
fixedtomovingmat * fixpos. Both, fixpos and movpos are
4-component vectors in homogeneous coordinates in c-style
order (level,row,column,1)^T in micrometer.
gray_gamma=... (default: 0.5)
gamma correction of gray values before registration. Gray
values are mapped by newgray = oldgray^gamma. Usually it is
a good idea to use a gamma value of 0.5 for images with
Poisson noise, because it approximately maps it to constant
gaussian noise.
interpolation=... (default: trilinear)
gray value interpolation method when transforming the
images. Can be 'trilinear' or 'nearestNeighbour'. Use
trilinear interpolation for real images and
nearestNeighbour for indexed images (e.g. segmentation
masks).
extrapolation=... (default: none)
Extrapolation method for the deformation field outside the
mask. Use 'none' for no extrapolation or 'TPS' for thin
plate spline extrapolation (can be very slow!) and 'fast'
for fast incremental extrapolation.
process_element_size_um=... (default: 2)
voxel-size for registration process in micrometer.
Registration needs cubic voxels.
padding_um=... (default: 80,80,80)
size of padding borders (levels, rows, columns) in
micrometer. The padding borders will be added to the fixed
image. The resulting image must be large enough that the
relevant parts of the moving image (after initial
transformation) fit into it.
lambda=... (default: 2)
lambda specifies the relative weight between data and
smoothness term during registration. E.g. lambda=2 adjusts
the internal weights, such that the maximal elongation is 2
and the maximal shortening is 1/2. These extreme
deformations may only appear, if the similarity measure
(normalized cross correlation) "insists" on this position
(by a jump from 0 to 1). So usually, the obtained
deformation is much smaller
gridSpacing_pix=... (default: 10,10)
Spacing of control points [pixel] in each iteration. e.g.
'10,10,5' would result in a grid spaciing of 10 pixels in
the first and second iteration and a grid spaceing fo 5
pixels in the third iteration
patchRadius_pix=... (default: 5,5)
patch radius [pixel] in each iteration. 'Patch' denotes is
the small image at each control point, that is used for the
normalized cross correlation
labelsSpacing_pix=... (default: 2,1)
spacing of labels [pixel] in each iteration. The 'labels'
denote the displacment hypotheses for each patch. The
hypotheses are placed along rays in axis and in diagonal
diretions.
labelsRadius_npt=... (default: 20,20)
number of labels (displacement hypotheses) along each ray
in each iteration.
debuglevel=... (default: 0)
Level of extra debugging output:
0: none
1: extra file with intermediate images 'debug.h5'
2: also results from ncc in 'debug.h5'
3: very verbose during optimization